Principal Investigators
Core unit “Genome signatures and integrated systems biology of pathogen-host interaction and cellular networks in the infected and injured lung”
Summary
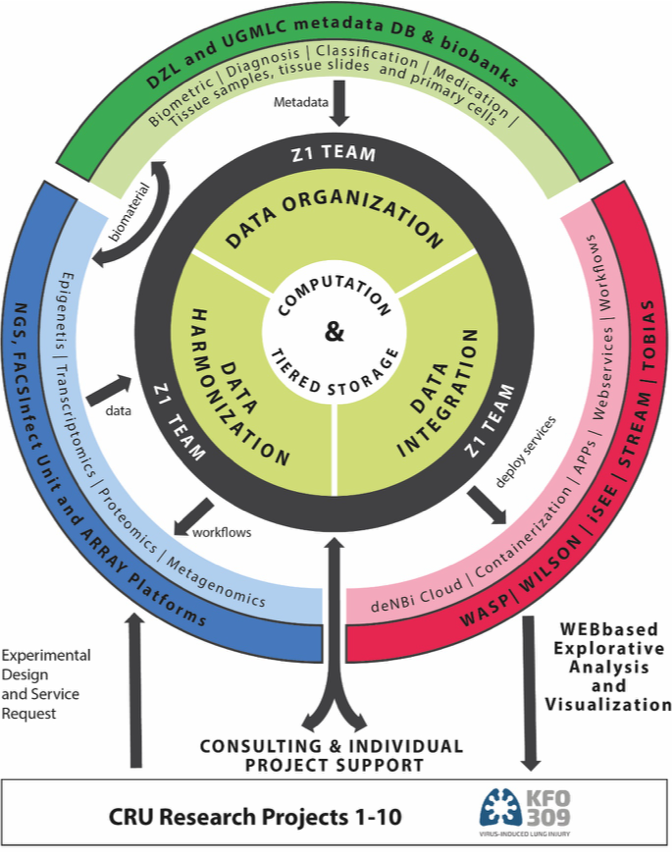
Understanding cell-specific gene regulatory events and their impact on cellular networks in infected lungs requires an integrative CRU platform which delivers high-throughput transcriptomics, genomics and epigenetics analyses from primary lung cells/organoids/tissues including highly purified cell populations even of rare subsets, as well as advanced tools for data exploitation. The core unit (Z1) provides methods and tools for processing of murine and human lung specimens including patient samples to all CRU projects, with a focus on cell specificity and after prior processing/enrichment of rare cell populations by Auto-MACS and FACS. Furthermore, automated sample/library preparation, whole-transcriptome analyses by microarrays and next-generation sequencing (NGS) including single cell RNA sequencing (scRNA-Seq) and droplet-based approaches (Drop-Seq) for transcriptomics and epigenomics, as well as microbiome/metagenome analysis are cutting edge techniques provided by Z1. A comprehensive portfolio of bioinformatics software solutions is offered to all CRU members to process and analyze the above-mentioned data sets in a standardized and highly automated fashion.